CRISPR are repeat arrays found in the DNA of many bacteria and archaea.
The name is an acronym for Clustered Regularly Interspaced Short Palindromic Repeats.
The repeats or DR, ranging in size from 23 to 47 base pairs, are separated by spacers of similar length.
Repeats often show some dyad symmetry but are not truly palindromic. Spacers are usually unique in a genome.
They match sequences in genomes of phage, plasmid or mobile genetic elements. Inside a species, the CRISPR repeat array may show polymorphism.
Cas genes stand for CRISPR-associated genes. Together with the CRISPR array they constitute the CRISPR-Cas defense mechanism. Cas function as clusters of 3 to more than 10 genes and can be distributed into 6 types (I to VI) and more than 30 subtypes.
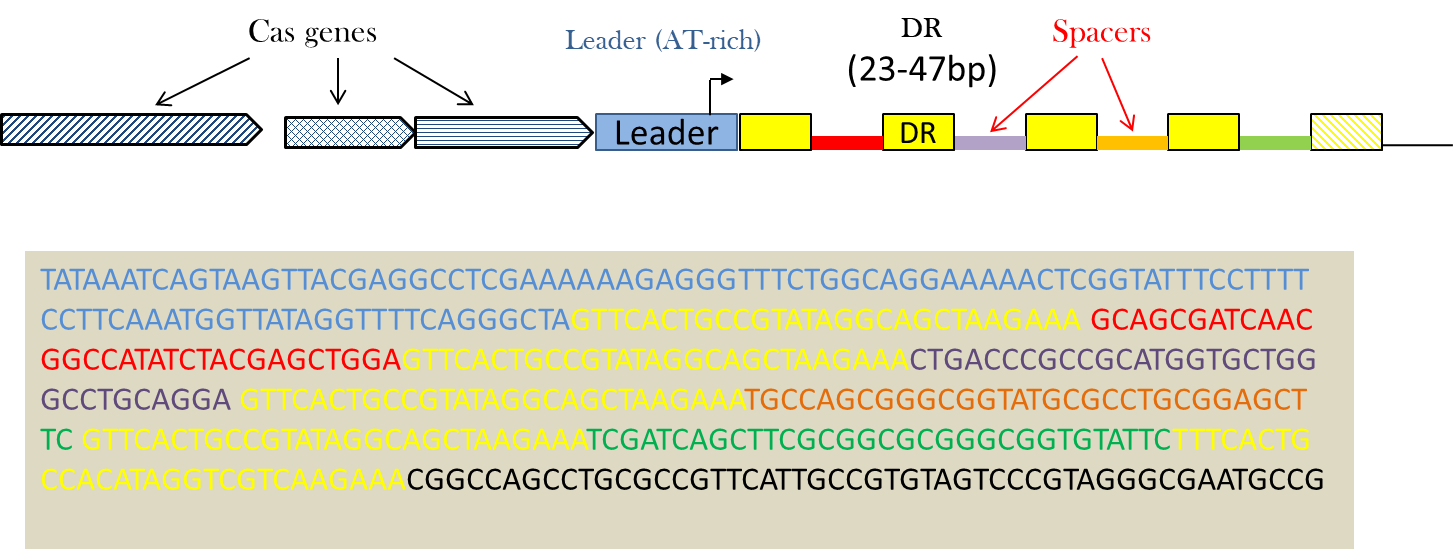
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) are specific structures found in many prokaryotic genomes that show characteristics of both tandem and interspaced repeats. They have been described in a wide range of prokaryotes, including the majority of Archae and many Eubacteria. A CRISPR locus is characterized by:
In a given strain several CRISPRs can be found with a single or different repeat sequences but only one of each kind is associated
with the cas genes. The spacers in the different CRISPRs are different.
The unique sequences or spacers correspond mostly to fragments of foreign DNA, ie. viruses, plasmids or mobile genetic elements.
Several genes called cas for CRISPR-associated are found in the vicinity of CRISPRs and perform the three different functions of the immune system: adaptation, crRNA maturation and interference. Their number varies from one type to another. Phylogenetic studies performed on the CAS protein suggest that CRISPRs are acquired by horizontal transfer. This is further shown by their presence on megaplasmids.
CRISPR loci are transcribed into a pre-crRNA from the leader acting as a promoter. This precursor is then matured into small crRNA that play a role in the targeting and destruction of homologous foreign sequences.