Search
A multi-criteria search can be performed by :
- Strain name (ex : "Actinoalloteichus hoggarensis"), or assembly/sequence accession (ex : "GCA_002234535.1")
- Kingdom (either “All” prokaryotes, or selectively Archaea or Bacteria)
- CRISPR/Cas element to be displayed in the table.
- Taxonomy Id (ex:2070 to display all Pseudonocardiaceae family, or 1470176 for Actinoalloteichus hoggarensis species).
- CAS genes
- CAS cluster type
1- Release
The date of the sequence release is indicated, and can be sorted by clicking on the column header.
2- Sequences: SEQ
The database was created from sequences associated with complete Genbank genomes and chromosomes.
A strain can contain one or several chromosomes and plasmid sequences, referred as sequence or SEQ,
with its own identification number.
In the following example one chromosome and two plasmids are present in the strain:
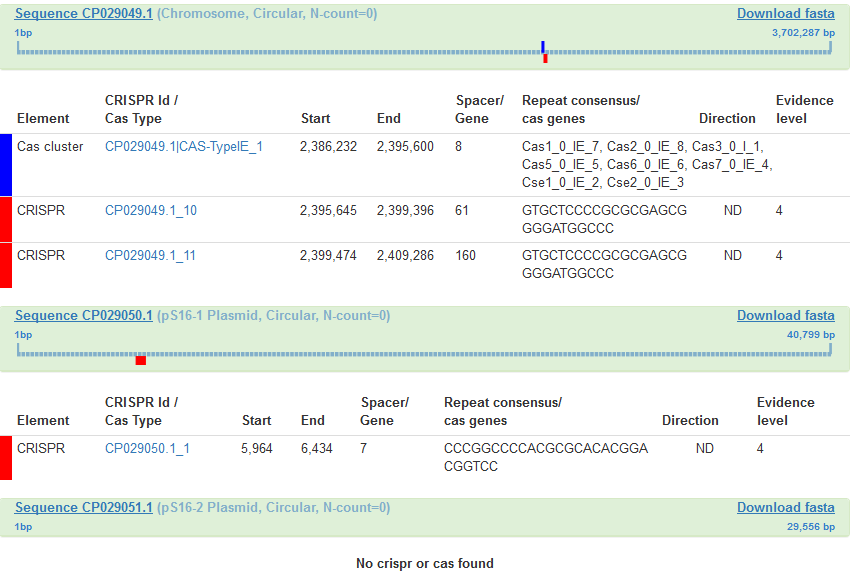
3- CAS
The number of cas gene clusters is indicated.
Cas proteins are identified by HMM search following annotation of ORFs by Prodigal. HMM profiles were deduced
from analysis of reference Cas proteins [Haft, 2001 #2354;Haft, 2005 #720].
Clusters of Cas proteins can be assigned to type and subtype according to the presence or absence of particular “signature”
genes and the architecture of the cas operon [Makarova, 2015 #2248] [Shmakov, 2015 #2249] [Abby, 2014 #2156]. cas3/cas7,
cas9, cas10, cas12 and cas13 have been defined as the signature genes for type I, type II, type III, type V and type VI respectively.
To build the database, the sub-type model of the program CasFinder version 2.0 was used. Consequently, incomplete clusters
were not considered and not shown in the database.
4- CRISPR
The number of CRISPR arrays is indicated.
CRISPR arrays are identified using CRISPRFinder and given an evidence level based on the number of repeats,
their internal conservation, and the % of similarity between spacers. Level 4 CRISPRs have the best score. Level 1
and level 2 arrays have little chance to be real CRISPR arrays. Level 1 CRISPRs which contain less than three spacers
are upgraded to level 4 when a similar DR is found in a level 4 CRISPR.
Results
1-Sequence(s)
The Elements detected in each sequence of the selected strain are displayed with their positions on the genome and relevant characteristics. Download fasta allows recovering the sequence in a fasta format.
Activating an element leads to details and allows recovering sequences in fasta format or their reverse complement. Repeats and spacers can be Blasted against lists of repeats and spacers present in the database by selecting the sequence of interest and starting the search.
2-Assembly info
This gives access to information concerning the sequence retrieved from Genbank.
3-Taxonomy
Provides the ordered taxonomy of the selected strain.
4-Analyzer
It gives the version of CRISPRCasFinder used to perform the analysis, the date of analysis, and command line for CRISPRCasFinder program.
